IPC Progress
➢ IPC has developed a new bioinformatic approach of identifying specific pathogen peptide sequences that represent an enhanced T-Cell proliferation phenotype.
➢ IPC has filed patents on the bioinformatics technology and pathogen specific sequence patterns.
➢ IPC is currently testing the patterns we have identified from Malaria, HIV-1, MS, MG & TB for establishing antigenic responses in in-vitro tests.
➢ Utilizing this new method IPC has generated a comprehensive list of pathogen specific PAMPs for a variety of viruses, bacteria and parasites for vaccine application testing.

DCs harvested from Normal Donor samples and matured with specific PAMP then reintroduced to the Donor sample after maturation to observe T-Cell proliferation.
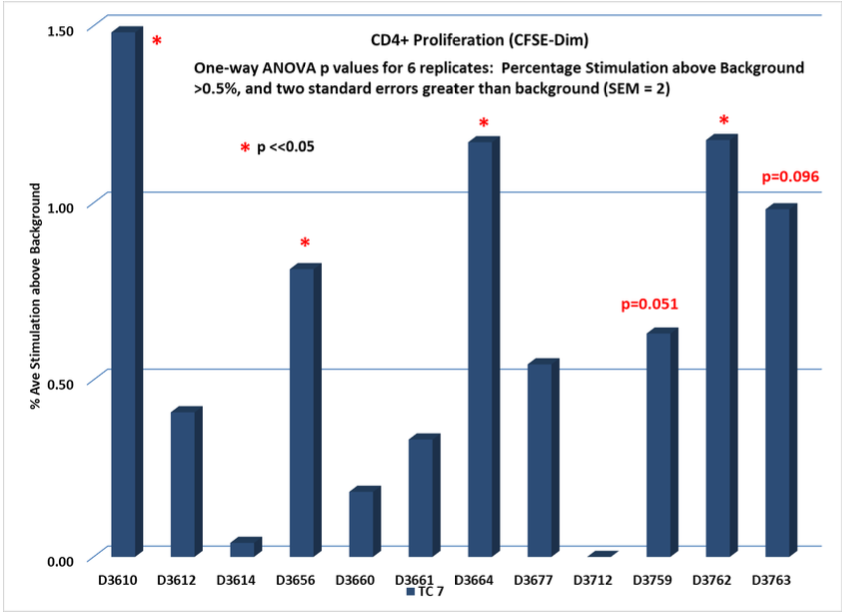
Test Condition 7 (TC 7)
MHC I + MHC II PAMP identified in TB Env protein.
Single PAMP tested against 12 random normal donors measuring induced antigenicity
Classical CD4+ Response

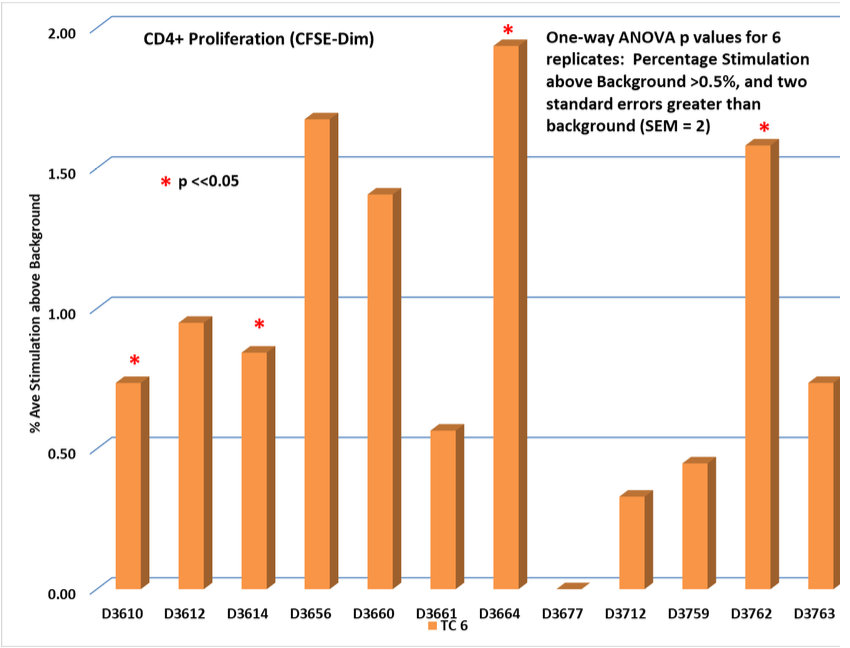
Test Condition 6 (TC 6): MHC I + MHC II PAMP PAMP identified in HIV-1 gp 41.
PAMP tested against 12 random normal donors measuring induced antigenicity.
Classical CD4+ Response

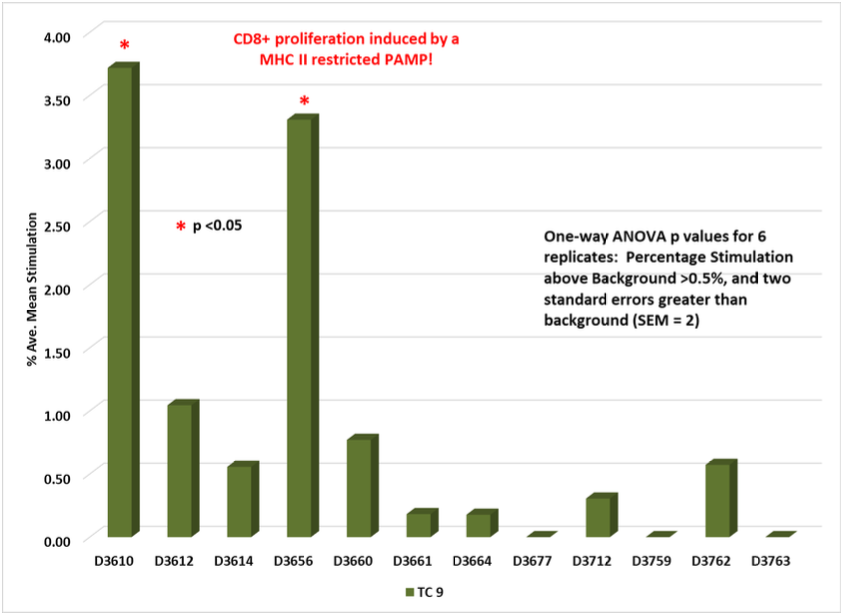
Test Condition 10 (TC 10):
MHC I PAMP identified in HIV-1 gp 41.
PAMP tested against 12 random normal donors measuring induced antigenicity.
Non-Classical CD8+ T-Cell Response

IPC BioInformatics for ETCP PAMP Identification
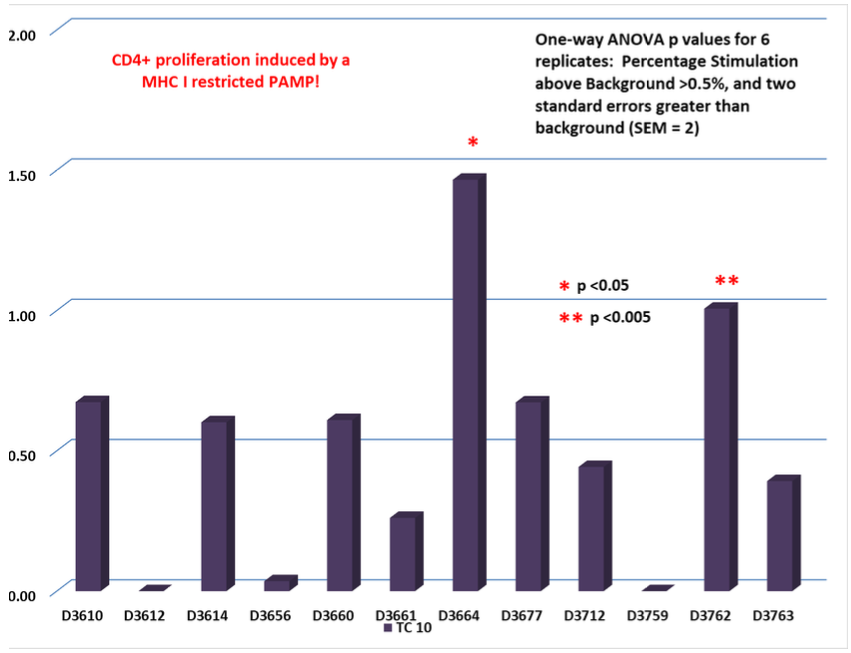
Test Condition 10 (TC 10):
MHC I PAMP Identified in HIV-1 gp 41.
PAMP tested against 12 random normal donors measuring induced antigenicity.
Non-Classical CD4+ T-Cell response
